Qinghua Wanga,†, Tingting Dua,†, Zhihui Zhanga, Qingyang Zhanga, Jie Zhangb,Wenbin Lic, Jian-Dong Jianga, Xiaoguang Chena,*, Hai-Yu Hua,*
a State Key Laboratory of Bioactive Substances and Function of Natural Medicine, Beijing Key Laboratory of Active Substances Discovery and Drugability Evaluation, Institute of Materia Medica, Chinese Academy of Medical Sciences & Peking Union
Medical College, Beijing 100050, China
b Sichuan Jiuzhang Biological Science and Technology Co., Ltd., Chengdu 610041, China
c Cancer Center, Beijing Tiantan Hospital, Capital Medical University, Beijing 100070, China
†These authors made equal contributions to this work.
*Corresponding authors.
Developing a photo-affinity probe PAL-CGA revealed ACAT1 as CGA’s key target,and demonstrated CGA inhibits cancer proliferation by blocking ACAT1’s Y407 phosphorylation, highlighting its clinical potential in cancer therapy.
Abstract Chlorogenic acid (CGA) is a natural product that effectively inhibits tumor growth, demonstrated in many preclinical models, and phase II clinical trials for patients with glioma. However, its direct proteomic targets and anticancer molecular mechanisms remain unknown. Herein, we developed a novel bi-functional photoaffinity probe PAL/CGA and discovered mitochondrial acetyl-CoA acetyltransferase 1 (ACAT1) was one of the main target proteins of CGA by using affinity-based protein profiling (AfBPP) chemical proteomic approach. We performed in-depth studies on ACAT1/CGA interactions via multiple assays including SPR, ITC, and cryo-EM. Importantly, we demonstrated that CGA impaired cancer cell proliferation by inhibiting the phosphorylation of tetrameric ACAT1 on Y407 residue through a novel mode of action in vitro and in vivo. Our study highlights the use of AfBPP platforms in uncovering unique druggable modalities accessed by natural products. And identifying the molecular target of CGA sheds light on the future clinical application of CGA for cancer therapy.
KEYWORDS: AfBPP; Natural product; Chlorogenic acid; Probe; ACAT1
1. Introduction
Chlorogenic acid (CGA), an ester formed from caffeic acid and L-quinic acid, is a natural product often found with high concentrations in green coffee beans1,2. CGA possesses a large spectrum of biological activities such as antidiabetic-, DNA- or neuroprotective effects3-5. Since the past few years, with the great improvement in the industrial scale of preparation and purification of CGA from raw plant extracts, CGA has become a novel agent, which has not only been tested in preclinical models but also in clinical trials for solid tumors in China (phase I, NCT02728349, Apr. 2016)6. A phase II study is ongoing focusing on malignant glioblastoma (NCT03758014, Nov. 2018). Mechanistically, an increasing number of studies show that CGA functions through multiple layers of cellular regulation, including suppression of inflammation, cellular senescence, epithelial-mesenchymal transition (EMT), genetic alterations, TGF-β overproduction, induction of tumor cell apoptosis, extra-cellular matrix modification7-11. However, the direct proteomic target of CGA remains unknown.
Activity-based protein profiling (ABPP) is one of a growing number chemical proteomic approaches that combines activity-based probe (ABP) and proteomics technologies to identify protein targets of bioactive natural products to help elucidate their mode of action and side effects12-15. The comprehensive fishing and identification of protein target(s) consist of three key steps: (1) probe design and synthesis, (2) target fishing and protein identification, (3) target confirmation and validation. When natural products of interest can covalently modify their targets, a tag can be engineered for target enrichment16,17. However, many natural products bind non-covalently to their tentative targets or substrates18,19. ABPP with affinity-based probes (AfBPs) can be used to study the interactions between reversibly binding natural products and their targets20-22. AfBPs derived from non-covalent bioactive compounds require the installation of photo-crosslinking linkers. Upon ultraviolet (UV) irradiation, the photoreactive crosslinking group generates a highly reactive intermediate that reacts with its adjacent molecule, resulting in the formation of a covalent bond between the probe and the target protein23,24. A major challenge in developing such probes is the interference with bioactivity when modifications are introduced to the parental compound’s scaffold25,26.
This study aims to elucidate the protein target of CGA. To achieve this, we developed a novel AfBP, PAL/CGA, which incorporated photoaffinity and enrichment handles, based on the structure‒activity relationship (SAR) of CGA. We employed proteomics approaches to discover and identify the direct protein target of PAL/CGA. To comprehensively investigate the interaction between CGA and its target protein, we systematically examined the binding affinity, binding site, and mode of actions of CGA with its protein target through biochemical and structural biology techniques such as surface plasmon resonance (SPR), isothermal titration calorimetry (ITC), and cryoelectron microscopy (cryo-EM). Further, we dissected CGA’s mechanism of action in cell based- and xenograft animal models.
2. Methods
2.1. Materials and reagents
All the chemicals were purchased from J&K. Commercially available reagents were used without further purification. CGA was obtained from Sichuan Jiuzhang Biological Science and Technology Co., Ltd. Point mutations were introduced by using quikchange XL site-directed mutagenesis kit (Agilent Technologies). Isolation of intact mitochondria from A375 tumor tissues and A375 cells was performed using a mitochondria isolation kit (Beyotime). ShRNA construct for human ACAT1 knockdown was from Open Biosystems. The two shRNA sequences for ACAT1 knockdown are:
5′-GCCACTAAGCTTGGTTCCATTCTC-3′ (shRNA#1) and
5′-GCCTTTAGTCTGGTTGTACTACTC-3′ (shRNA#2). Fluorescence emission spectra and full wavelength absorption spectra were performed on Biotek Synergy™ H1. Confocal laser scanning microscope imaging was performed using a Leica TCS SP2 Confocal Microscope. 1H NMR and 13C NMR spectra were recorded at 500 MHz and 125 MHz, respectively. HRMS was assessed utilizing a Thermo LCQ Deca XP Max mass spectrometer equipped with ESI.
2.2. Synthetic procedures and characterized data of CGA/PAL compound
CGA (200 mg, 0.56 mmol), carboxymethylcellulose sodium (CMC) (136 mg, 0.56 mmol), 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride (EDCI) (130 mg, 0.67 mmol) and N-hydroxybenzotrizole (HOBT) (92 mg, 0.67 mmol) were dissolved in dry tetrahydrofuran (THF) (2 mL) and stirred in an ice-water bath for 30 min. After then, 2-(3-(but-3-yn-1-yl)-3H-diazirin-3-yl) ethan-1-amine (77 mg, 0.56 mmol) was added, and the reaction mixture was stirred for 4 h. The reaction mixture was removed under reduced pressure and the product was purified by preparative HPLC (H2O + 0.05% TFA; 5%‒95% CH3CN + 0.05 % TFA), and got white solid 78 mg, yield 76%. 1H NMR (500 MHz, Methanol-d4) δ 8.03 (t, J = 6.2 Hz, 1H), 7.59 (d, J = 15.9 Hz, 1H), 7.07 (s, 1H), 6.96 (d, J = 8.1 Hz, 1H), 6.80 (d, J = 8.0 Hz, 1H), 6.31 (d, J = 15.9 Hz, 1H), 5.43 (td, J = 10.9, 4.8 Hz, 1H), 4.27 (s, 1H), 3.74 (d, J = 9.7 Hz, 1H), 3.14 (q, J = 6.1 Hz, 2H), 2.28 (s, 1H), 2.17 (d, J = 11.0 Hz, 1H), 2.09 (t, J = 13.3 Hz, 1H), 2.02 (s, 4H), 1.64 (s, 4H); 13C NMR (125 MHz, Methanol-d4) δ 176.7, 176.6, 169.0, 149.5, 147.0, 146.7, 127.7, 122.9, 116.5, 115.3, 115.1, 83.6, 77.7, 74.3, 72.6, 71.9, 70.4, 39.9, 38.6, 35.3, 33.1, 27.8, 13.8; HRMS (m/z) [M+H+]: calcd. for C23H28N3O8 +: 474.1871; found: 474.1871.
2.3. Photoaffinity purification24
CGA was used as a control to compare with PAL/CGA, a photoaffinity labeling probe. CGA was incubated with mitochondrial proteins at 50 μmol/L, while PAL/CGA was added at 10 μmol/L, both at 4 °C for 2 h. Subsequently, the samples were exposed to UV light (365 nm) at 4 °C for 30 min, followed by the addition of azide biotin at 20 μmol/L. A catalyst, composed of CuSO4 (20 μmol/L) and TECP (25 μmol/L), was used to reduce Cu(II) to Cu(I), and THPTA (60 μmol/L) was added to stabilize Cu(I). After 18 h of rotation at room temperature, proteins were precipitated with cold acetone, washed thrice, and dissolved in 1% SDS in PBS. PAL/CGA-bound proteins, isolated using streptavidin beads (Invitrogen), were eluted, subjected to SDS-PAGE, and visualized with coomassie blue staining. Bands differing in abundance between the CGA-preincubated and control samples were analyzed by LC‒MS/MS using a Q Exactive Mass Spectrometer (Beijing Protein Innovation, China). Protein identification was performed using Mascot Daemon (version 2.3.0).
2.4. Cell culture
A375 cells (ATCC® CRL-1619™, human melanoma) and NCI-H1299 cells (ATCC CRL-5803, human non-small cell lung carcinoma) were acquired from the China Infrastructure of Cell Line Resources, Beijing, China. The A375 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Corning), supplemented with 10% fetal bovine serum (FBS) (Invitrogen) and 1% penicillin–streptomycin (Corning). The culture conditions were maintained at 37 °C under two different oxygen levels: normoxia (95% air, 5% CO2) and hypoxia (1% O2, 94% N2, and 5% CO2). Similarly, NCI-H1299 cells were cultured in Roswell Park Memorial Institute 1640’s medium (RPMI 1640, Corning), also supplemented with 10% FBS (Invitrogen) and 1% penicillin–streptomycin, under the same normoxic and hypoxic conditions.
2.5. Cell viability assay
The cytotoxicity of CGA on A375 cells was assessed using the MTT assay. Cells were seeded at 1×104 cells/mL in 96-well plates, incubated at 37 °C in 5% CO2 for 12 h under hypoxic conditions. CGA was then applied in concentrations of 1.25–10 μmol/L for 72 h under both conditions. Post-treatment, MTT solution (5 mg/mL in PBS) was added, and after 4 h, replaced with 150 μL DMSO per well. After another 4 h, absorbance was measured at 490 nm using a Biotek Synergy™ H1. Cell viability was calculated using Eq. (1): Cell viability (%) = (A‒A0) /(As‒A0) ×100 (1) where A is the absorbance of the experimental group, As of the control, and A0 of the blank. The experiment was replicated thrice.
2.6. Recombinant protein purification
The pET28_ACAT1_His plasmid, provided by Cheryl Arrowsmith (Addgene plasmid #25510; http://n2t.net/addgene:25510; RRID: Addgene_25510), was used for ACAT1 protein expression in E. coli, induced overnight with isopropyl-D thiogalactopyranoside (IPTG). Post-induction, the cells were sonicated, and the lysate was centrifuged at 12,000 rpm for 30 min to remove cellular debris. The supernatant was then applied to a 5 mL HisTrap HP column (GE Healthcare) for affinity chromatography, followed by extensive washing to purify His-ACAT1 protein. The protein was desalted using a HiTrapTM Desalting column, further purified through a HiTrapTM Q HP anion exchange column and Sephadex G-200 size exclusion column (GE Healthcare), and concentrated to about 5 mg/mL using a 50 kDa concentrator. Protein concentration and purity were confirmed by SDS-PAGE.
2.7. Thermal shift assay (TSA)
Purified recombinant ACAT1 (40 μg/mL) was incubated with CGA (0.1, 1, 10 μmol/L) and AH (10, 20, 40 μmol/L) on ice for 1.5 h. The samples were then heated at various temperatures (47 to 52 °C) for 3 min and cooled at room temperature for 3 min. Subsequently, the samples were centrifuged at 2000 rpm for 20 min at 4 °C. The supernatants were analyzed by SDS-PAGE, followed by Western blot.
2.8. Drug affinity responsive target stability (DARTS) assay
Cells were harvested and lysed using a buffer containing 0.4% Triton X-100, 400 mmol/L NaCl, 100 mmol/L Tris-HCl (pH 7.5), and 20% glycerol to isolate total proteins. The lysates were diluted 1:10 with TNC buffer (50 mmol/L Tris-HCl, pH 8.0, 50 mmol/L NaCl, 10 mmol/L CaCl2) and treated with various concentrations of CGA or DMSO as a control for 1 h at room temperature. Pronase (25 μg/mL) was then added and incubated for 30 min at 37 °C. The reactions were stopped by adding SDS-PAGE loading buffer, and the samples were analyzed via Western blot using an anti-ACAT1 antibody.
2.9. Western blot analysis
After treatments, cells were harvested, washed with cold PBS, and lysed for 30 min on ice using Solarbio lysis buffer. The lysates were centrifuged at 12,000 rpm for 30 min at 4℃, and protein concentrations in the supernatant were quantified using the Bradford assay (Yeasen). Proteins were resolved on 10% SDS-PAGE and transferred to PVDF membranes (Millipore) using a 10 mmol/L CAPS (pH 11.0, 10% methanol) transfer buffer. The membranes were blocked for 1 h at room temperature in 5% nonfat dry milk/TBST and incubated overnight at 4 °C with primary antibodies against p- ACAT1 (Signalway Antibody, #SAB491P), ACAT1 (Proteintech), and β-actin (Zhongshan Golden Bridge Biotechnology). After TBST washes, membranes were incubated with HRP-conjugated secondary antibodies (Zhongshan Golden Bridge Biotechnology) and developed using Western Bright ECL (TANON Science & Technology).
2.10. Cryo-EM single particle analysis of ACAT1/CGA complex
Preparation: Purified ACAT1 protein (0.9 mg/mL) was incubated with CGA (0.2 mmol/L) on ice for 1 h, then used to prepare cryo-EM grids or diluted 800-fold for negative staining. Cryo-EM procedure:
a) Sample preparation: Using a Thermo Fisher Vitrobot Mark IV, 3.5 μL of the ACAT1/CGA complex was applied to a glow-discharged holey carbon grid with reduced graphene oxide film. After blotting for 1.0 s at 100% humidity and 8 °C, the grid was flash-frozen in liquid ethane. b) Data collection: Data were collected on a Thermo Fisher Titan Krios G3i electron microscope with a Gatan K3 camera, at 300 kV. Images were taken at a magnification of 105 k, yielding a pixel size of 0.669 Å. A total of 2269 movie stacks with 32 sub-frames each were recorded, using EPU for a total dose of 50 electrons/Å2. c) Image processing: The image stacks were processed with MotionCor2 for alignment and summation, producing micrographs at a 0.669 Å pixel size. CTF estimation was done using CTFFIND4.1. Particle picking was automated in RELION3.1, followed by 2D and 3D classification, with the latter using a PDB-derived map for reference. Final 3D auto-refinement was performed with D2 symmetry, and resolution was determined by the gold-standard FSC 0.143 criterion.
2.11. Native PAGE
The oligomerization of ACAT1 was analyzed using discontinuous native gel electrophoresis. The 5 mL, 5% stacking gel consisted of 3.4 mL H2O, 0.5 mL 1 mol/L Tris-HCl (pH 6.8), 0.83 mL 30% acrylamide, 0.05 mL 10% ammonium persulfate, and 5 μL TEMED. The 10 mL, 8% separating gel was made of 4.6 mL H2O, 2.5 mL 1.5 mol/L Tris-HCl (pH 8.8), 2.7 mL 30% acrylamide, 0.1 mL 10% ammonium persulfate, and 10 μL TEMED. Samples were prepared with the NativePAGE™ Sample Prep Kit (Invitrogen). The anode buffer contained 50 mmol/L BisTris, 50 mmol/L Tricine (pH 6.8), and 0.002% Coomassie Brilliant Blue G-250. After electrophoresis, the gel was either scanned directly or used for Western blot analysis on a PVDF membrane.
2.12. ACAT1 activity assay
An in vitro ACAT1 activity assay was performed as described previously to identify CGA as ACAT1 inhibitors. In brief, 100 ng purified ACAT1 protein was added to assay buffer containing 50 mmol/L Tris-HCl (pH 8.1), 20 mmol/L MgCl2, 40 mmol/L KCl, 20 μmol/L acetoacetyl-CoA, and 60 μmol/L CoA. The change in absorbance at 303 nm was measured using a Biotek Synergy™ H1.
2.13. Surface plasmon resonance (SPR) assay
The interaction between ACAT1 and the compounds CGA or AH was assessed using the Biacore T200 system (GE Healthcare, Uppsala, Sweden) at 25 ℃. Recombinant human ACAT1 protein was immobilized on a CM5 sensor chip using an amine coupling kit (GE Healthcare, Buckinghamshire, UK), achieving immobilization levels of approximately 15,000 RU (Response Units). The compounds were then injected as analytes at various concentrations into a 25 mmol/L Tris-HCl buffer (pH 7.5), serving as the running buffer. Binding affinity studies involved injecting analytes at specified concentrations with a flow rate of 30 μL/min, allowing 120 s each for contact and dissociation phases. The chip was subsequently washed with the running buffer between analyses.
2.14. Isothermal titration calorimetry (ITC)
The binding interactions of CGA and AH with both wild-type ACAT1 and the ACAT1 A401F mutant were analyzed using an EAQ-ITC instrument (MicroCal, GE Healthcare, USA) at 25 ℃. CGA was prepared in a buffer containing 25 mmol/L Tris (pH 7.5) and 100 mmol/L NaCl. In the assay, 2 μL aliquots of ACAT1 (40 μmol/L) were injected at 150-s intervals into the ITC cell containing either 1.0 μmol/L CGA or 40 μmol/L AH. The resulting data were processed using the Origin 7.0 software, fitting to a two-site binding model to determine thermodynamic parameters such as enthalpy change (ΔH), entropy change (ΔS), and the equilibrium dissociation constant (KD).
2.15. Confocal imaging of cancer cells treated with CGA
Cancer cells, upon reaching 80% confluence, were seeded at 1 × 105 cells/mL (0.3 mL) into 8-well chamber slides with sterile coverslips. They were cultured for 24 h in DMEM or RPMI 1640 containing 10% FBS in a 5% CO2 humidified atmosphere at 37 ℃. After 30 min of treatment with 10 μmol/L CGA, the media were removed and cells were washed twice with PBS. Imaging was conducted using a Leica TCS SP8 X Confocal Microscope with a 63 × oil immersion objective, employing a high-pressure He-Ne lamp and diode laser for excitation at λex = 405 nm and λem = 430‒530 nm.
2.16. Pyruvate dehydrogenase activity assay
A375 cells were cultured in 100 mm dishes at 2×104 cells/mL and incubated in 5% CO2 at 37 ℃ for 12 h. The cells were then treated with CGA at concentrations of 0, 0.1, 1.0, and 5.0 μmol/L for 24 h. Mitochondria were isolated from these cells using a Beyotime mitochondria isolation kit. The activity of pyruvate dehydrogenase in the mitochondria was measured using a Sigma‒Aldrich assay kit, following the manufacturer’s protocol.
2.17. Tumor xenograft model studies
Male athymic BALB/c-nu mice (8–10 weeks old, 20 g) were obtained from HFK BIOSCIENCE Co., Ltd., Beijing, China. Cancer cells, including A375 cells and NCIH1299 cells, both with and without ACAT1 knockdown, were prepared at 5 × 107–6 × 107/mL in PBS and injected subcutaneously into the right flank of the mice. Once tumors reached 100 mm3, mice were divided into groups and received daily intraperitoneal injections of either a saline vehicle (0.9% NaCl) or CGA (50 mg/kg). Tumor measurements were taken every three days. Mice were euthanized one hour after the last treatment, and tumor and body weights were recorded and analyzed using GraphPad Prism 8. This study was approved by the Ethics Committee for Animal Experiments of the Institute of Materia Medica, Chinese Academy of Medical Sciences & Peking Union Medical College, and complied with their animal experiment guidelines (No.00004201).
2.18. Immunohistochemical staining
For the immunohistochemical analysis, tumor tissues were fixed and subsequently embedded in paraffin prior to sectioning. These sections were incubated overnight at 4 ℃ with an anti-phospho-ACAT1 antibody (Signalway Antibody). Following incubation, sections were stained using a DAB Quanto kit (Golden Bridge Biological Technology, Beijing, China) and subsequently photographed for analysis.
2.19. Statistical analysis
Experimental results are presented as mean ± standard deviation (SD), with each experiment conducted in triplicate. Statistical significance was determined using oneway or two-way ANOVA for P value analysis. IC50 values were calculated using nonlinear regression in GraphPad Prism 8. Graphical representations also display data as mean ± SD. Statistical significance is denoted as *P < 0.05, **P < 0.01, and ***P < 0.001, indicating increasing levels of significance.
3. Results and discussion
3.1. Design and synthesis of the novel photoaffinity labeling CGA analogue, PAL/CGA
The design of the AfBP must fulfill several prerequisites: First, a desirable synthetic strategy that introduces modifications at accessible entry points. Second, derivatization at this site does not affect the activity of parent compound CGA. Third, the corresponding probes should be detached from any bulky marker that would affect its interaction with targets during protein labeling. Based on SAR studies of CGA analogues, we inferred that the carboxylic acid moiety on the quinine acid could be modified without loss of CGA activity27-29. Since installation of the bulky biotin might significantly affect the activity and cell permeability of the probe, the bio-orthogonal approach was applied. Upon covalent attachment to its targets after UV irradiation, the reporter group is then introduced to the labeled targets via bio-orthogonal reaction30-32. To minimize the influence of the bioactivity of the probe, a bifunctional photo-crosslinker containing an aliphatic diazirine group and a terminal alkyne group in combination was selected and introduced to the carboxylic acid moiety of CGA via amide bond formation and yielded the photoaffinity labeling CGA analogue, PAL/CGA. The alkyl diazirine which is compact in size serves as the photo-reactive functional group. It generates the carbene intermediate upon UV irradiation and rapidly forms a covalent bond with target proteins. The terminal alkyne handle is used for the modification with marker biotin-azides after protein labeling via the click chemistry reaction, which allows proteins to be pulled down and identified by proteomic methods. The structure of probe PAL/CGA was characterized using 1H NMR, 13C NMR and HRMS (Scheme 1 and Supporting Information Scheme S1).
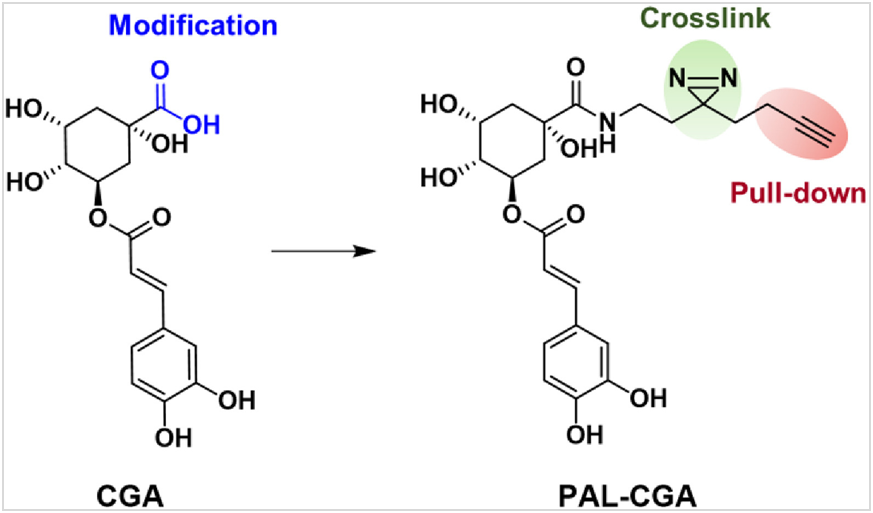
Scheme 1 Structures of CGA and PAL/CGA.
3.2. Effects of PAL/CGA on human melanoma cancer phenotypes
Our previous studies revealed that the natural product CGA decreased human melanoma A375 cell proliferation under hypoxia, but not normoxia conditions33. To assess the potency of the synthesized probe PAL/CGA, we monitored its inhibition of A375 cells under hypoxic conditions by MTT analysis. Consistent with the anti-cancer activity of CGA, PAL/CGA dose-dependently inhibited the proliferation of A375 cells at concentrations ranging from 2.5 to 10 μmol/L (Fig. 1a, Supporting Information Fig. S1), comparable to those for CGA, suggesting that the addition of the bifunctional tag did not noticeably interfere with the parental drug activity.
3.3. AfBPP to map CGA targets
We next applied the AfBPP chemo-proteomic platform to determine the specific protein targets of CGA. Based on colocalization imaging, we previously identified that CGA enriched in the mitochondria of cancer cells33. Therefore, mitochondria from A375 tumor tissues from xenograft models were extracted and isolated with a commercial mitochondria isolation kit (Supporting Information Fig. S2) and incubated with probe PAL/CGA, with or without CGA, followed by UV irradiation to produce the reactive species for crosslinking with target receptors. Subsequently, Cu(I) and biotin-azide were added, facilitating a click reaction to form a triazole ring between biotin and the PAL/CGA–protein complexes. These complexes were then isolated using streptavidincoupled Dynabeads and analyzed by SDS-polyacrylamide gel electrophoresis (PAGE). Fig. 1b shows that several protein bands appeared with PAL/CGA treatment, indicating their CGA specificity. Using liquid chromatography-mass spectrometry (LC–MS/MS) and Western blot analyses (Supporting Information Figs. S3 and S4, Table S1, Fig. 1c), we identified and confirmed that one of the fished proteins turned out to be mitochondrial ACAT1 (mitochondrial acetyl-CoA acetyltransferase 1). As shown in Fig. 1d and Supporting Information Fig. S5, CGA was potent in enzyme inhibition assays against purified recombinant ACAT1 (Supporting Information Fig. S6), exhibiting a half-maximal inhibitory concentration (IC50) value of 7.9 ± 0.34 μmol/L, a stronger effect than the positive control arecoline hydrobromide (AH), a reported covalent ACAT1 inhibitor tested together (Supporting Information Fig. S7). These results identified, for the first time, that ACAT1 is a novel direct target of CGA. We thus focused further characterization efforts on the interactions between CGA and ACAT1.
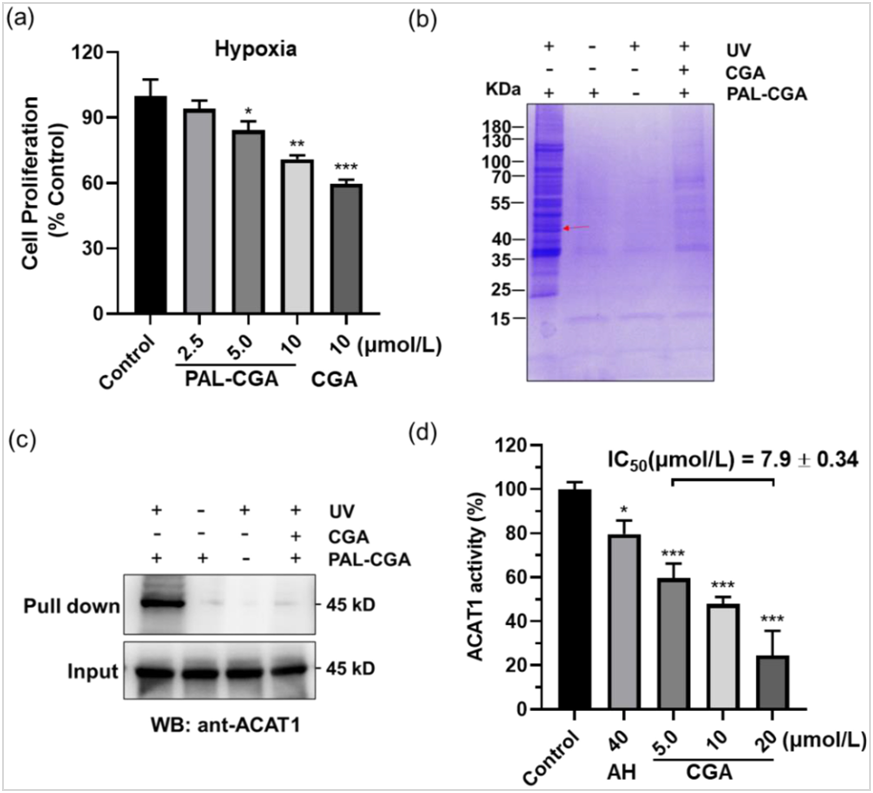
Figure 1 Identification of ACAT1 as the natural product CGA binding protein by a photoaffinity purification approach.
(a) PAL/CGA suppressed the proliferative ability of melanoma cancer cells; (b) Identification of ACAT1 as the binding protein of PAL/CGA (10 μmol/L) in mitochondria of the human cancer A375 tumors, which was competed off by CGA (50 μmol/L) preincubation. Coomassie blue staining of the PAL/CGA complex. The red arrow represented the position of ACAT1; (c) Validation of ACAT1 pulled down from mitochondria of the human cancer A375 tumors with PAL/CGA by Western blot; (d) Efficacy of CGA treatment in inhibition of purified recombinant ACAT1. IC50 values were determined. The bar graphs show the mean ± SD. * P< 0.05; **P< 0.01; ***P < 0.001.
3.4. Characterization of CGA interactions with ACAT1
Four label-free protein binding assays, including SPR, ITC, thermal shift assays (TSA), and drug affinity responsive target stability (DARTS), were set up to characterize the affinity and binding kinetics between CGA and ACAT1 (for details, see Methods section and Supporting Information). SPR analysis indicated the KD (equilibrium dissociation constant) value of CGA binding to ACAT1 was approximately 2.58 μmol/L (Fig. 2a). The ITC data was best fit using a two-site model. The first site in this model has a higher affinity (KD1 = 9.68 μmol/L) and larger enthalpy (ΔH1 = ‒51.8 kJ/mol). The second site exhibits weaker affinity (KD2 = 39.7 μmol/L) and a lower enthalpy change (ΔH2 = 3.35 kJ/mol). The ITC data suggested that interaction between GGA and ACAT1 results in complex formation characterized by non-covalent interactions (Fig. 2b).We next investigated whether CGA increased ACAT1 stability via a ligand‒protein complex in target engagement assays, with reference ACAT1 inhibitor AH as a positive control. From TSA, we found that CGA treatment efficiently protected ACAT1 protein from temperature-dependent degradation (Fig. 2c, Supporting Information Fig. S8). Meanwhile, DARTS assays demonstrated that CGA prevented pronase-induced proteolysis of ACAT1, which was CGA concentration-dependent. Interestingly, CGA was more effective in protecting ACAT1 proteolysis in lower concentrations in comparison with reference compound AH (Fig. 2d, Supporting Information Fig. S9).
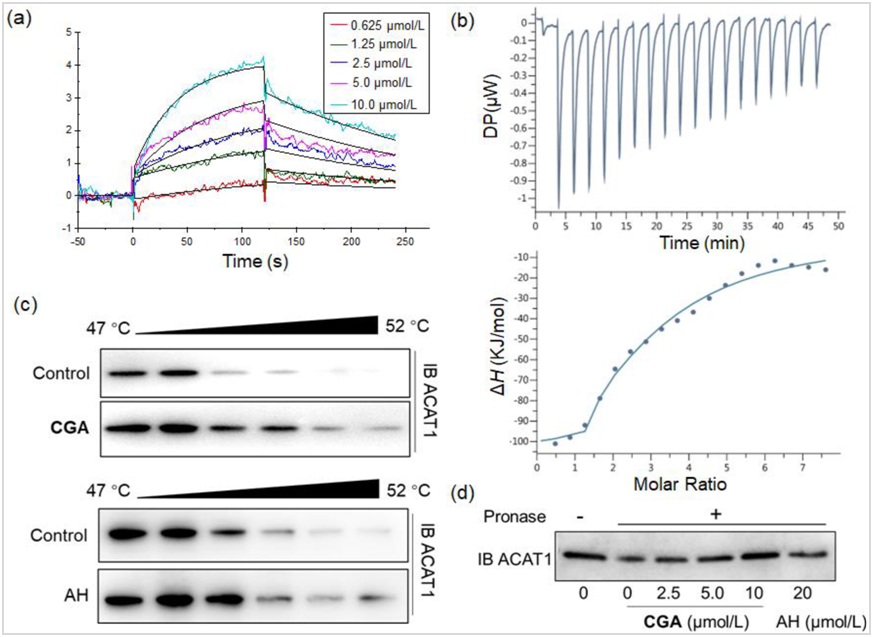
Figure 2 Characterization of CGA interactions with ACAT1 in vitro.
(a) Characterization of the binding affinity between CGA and ACAT1 by a SPR assay; (b) Binding affinity of CGA to ACAT1 determined by ITC; (c) Thermal shift assay using purified recombinant ACAT1, which were exposed to CGA (1.0 μmol/L) and AH (40 μmol/L); (d) CGA promotes target protein ACAT1 resistant to proteases (DARTS); data are representative of three independent experiments.
We next performed two complementary experiments to evaluate ACAT1‒CGA interaction in cell lysates and living cells. Two cell lines, A375 cells and human nonsmall cell lung carcinoma cells (NCI-H1299) were used. First, consistent with the results described above using purified recombinant ACAT1, DARTS assays showed that CGA treatment efficiently protected ACAT1 protein from dose-dependent degradation in both A375 and NCI-H1299 cell lysates, with the effects from 2 μmol/L CGA comparable with those from 40 μmol/L AH (Supporting Information Fig. S10).
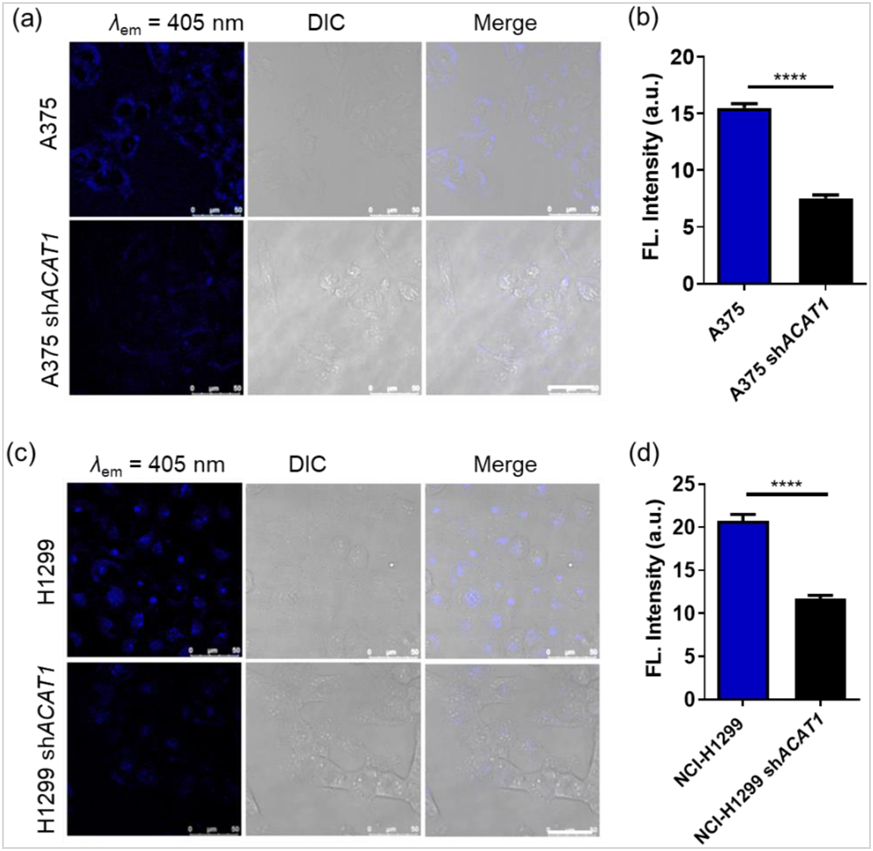
Figure 3 Identification of ACAT1 as a CGA-binding protein in vivo.
(a, c) Fluorescence images of A375 cells, NCI-H1299 cells and knockdown of endogenous ACAT1 cells incubated with CGA (10 μmol/L) for 2 h; (b, d) Quantification of fluorescence intensities from images above obtained for CGA in the A375 cells, NCI-H1299 cells and knockdown of endogenous ACAT1 cells. The bar graphs show the mean ± SD. *P< 0.05; **P < 0.01; ***P< 0.001.
Secondly, in our previous studies, CGA had shown high selectivity and robust staining intensity for mitochondria when it was used as a two-photon fluorescent probe33. With this feature, we examined whether the knockdown of ACAT1 led to a reduction of CGA labeling in mitochondria. A375 cells and NCI-H1299 cells were engineered with stable knockdown of endogenous ACAT1 by short hairpin RNA (shRNA) (Supporting Information Fig. S11). As shown in Fig. 3, the fluorescence intensities from CGA significantly decreased in ACAT1 knockdown cell lines (A375 shACAT1, NCI-H1299 shACAT1) compared with the fluorescence intensities in wildtype cells. All these experiments taken together provide evidence that ACAT1 is a specific CGA-binding protein.
3.5. Cryo-electron microscopy (EM) structure of ACAT1 in complex with inhibitor CGA
Previous studies showed that AH, the covalent inhibitor of ACAT1, bound to and disrupted ACAT1 tetramers into monomers, thus making tyrosine kinases unable to phosphorylate and stabilize active ACAT134. To elucidate CGA’s inhibitory mechanism, we initially attempted to crystalize the ACAT1/CGA complex without success. Consequently, we employed cryo-electron microscopy (cryo-EM) to investigate the structure of the ACAT1/CGA complex.
To identify the best sample for structure determination by cryo-EM, we firstly carried out negative-stain studies on full-length human ACAT1 in complex with CGA, confirming CGA’s non-disruptive effect on ACAT1 tetramer conformation through improved 2D class averages in the transmission electron microscopy (TEM) analysis (Fig. 4a, Supporting Information Fig. S12). With this sample, we accomplished cryo- EM reconstruction of the human ACAT1 homo-tetramer, reaching an overall resolution of 3 Å (Fig. 4b and c, Supporting Information Figs. S13, S14 and Table S2). In the structure of ACAT1 in complex with CGA, the inhibitor was stabilized by a hydrogen bond network, including the side chain of E234, E351, N377 and H397 (Fig. 4f). Interestingly, caffeic acid moiety of CGA was exposed to the outside of ACAT1 structure; whereas the quinic acid fragment of CGA was in an allosteric (Supporting Information Fig. S15 and Table S3). Catechol groups of caffeic acid moiety were next to the Y407 phosphorylation-binding site and interfered with the binding of phosphorylase to ACAT1 which would indirectly disrupt Y407 phosphorylation (Fig. 4d, e).
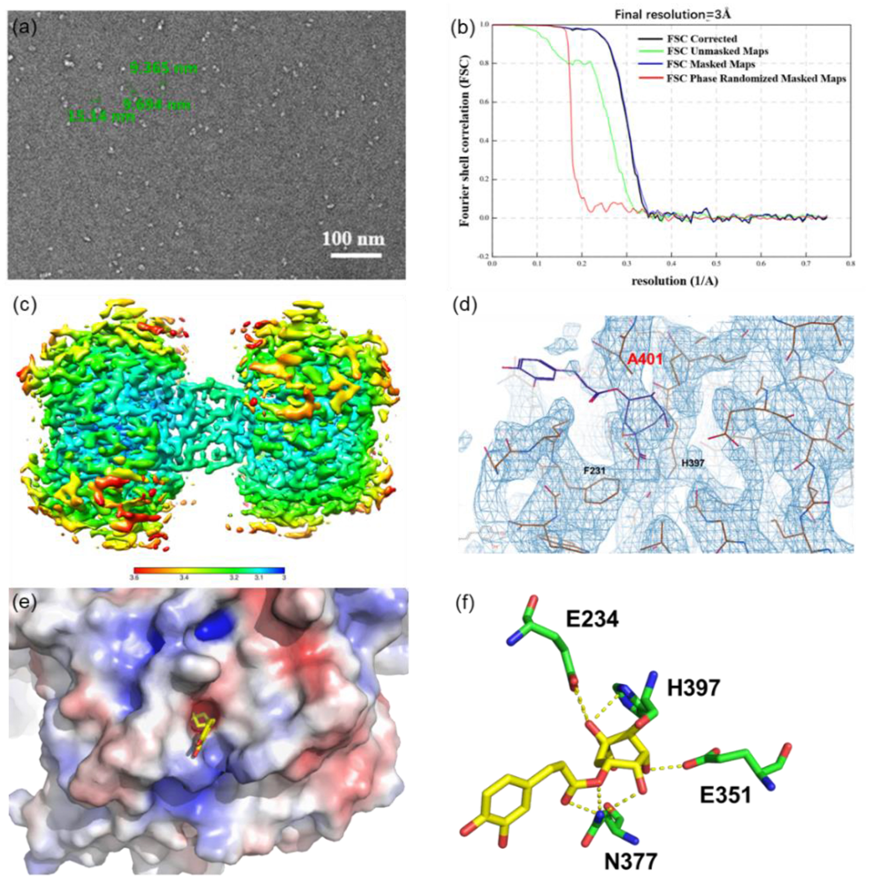
Figure 4 Cryo-EM structure of ACAT1 in complex with inhibitor CGA.
(a) Negative stain micrograph of ACAT1/CGA complex; (b) FSC curve of the map of ACAT1/CGA complex; (c) Local resolution of the map of ACAT1/CGA complex; (d) The superposition of cryo-EM structure of the ACAT1/CGA complex; (e) The crystallographic structure of the ACAT1/CGA complex; (f) Schematic drawing of the interactions between CGA and ACAT1.
To further confirm these results, we incubated purified tetrameric and monomeric ACAT1 proteins in the presence or absence of CGA for 12 h to assess whether CGA promoted disruption of ACAT1 tetramers on a native gel electrophoresis (Supporting Information Fig. S16). Contrary to the covalent inhibitor AH, there was no impact on the equilibrium between tetrameric and monomeric ACAT1 proteins when incubated with CGA, suggesting that the apparent activity inhibition of ACAT1 may be due to extensive conformational changes of tetramers, rather than disrupting ACAT1 tetramers into monomers.
To confirm that CGA directly targeted Ala 401 position in α-helical cavity of ACAT1, we perturbed position 401 by replacing Ala (A) with a large aromatic amino acid, Phe (F), which could prevent CGA binding to this site of ACAT1. Protein binding assays using ITC analysis further confirmed CGA was unable to bind with the mutant protein (Supporting Information Fig. S17). Overall, these analyses strongly supported the binding mode observed by cryo-EM structure, providing a clear view of the mode of action at the molecular level.
3.6. Effects of CGA on ACAT1 function
The deregulated metabolism is one of the hallmarks of cancer35-38. Recent studies have shown that ACAT1 is one of the important metabolism regulators for tumorigenesis39-43. Upregulated oncogenic tyrosine kinases phosphorylate the Y407 residue of ACAT1 and stabilize its active tetrameric form, which in turn inhibits pyruvate dehydrogenase (PDH) activity, promotes glycolysis, resulting in tumor growth34. Therefore, the inhibition of ACAT1 activity removes the inhibitory effect of ACAT1 on PDH and reduces tumor growth.
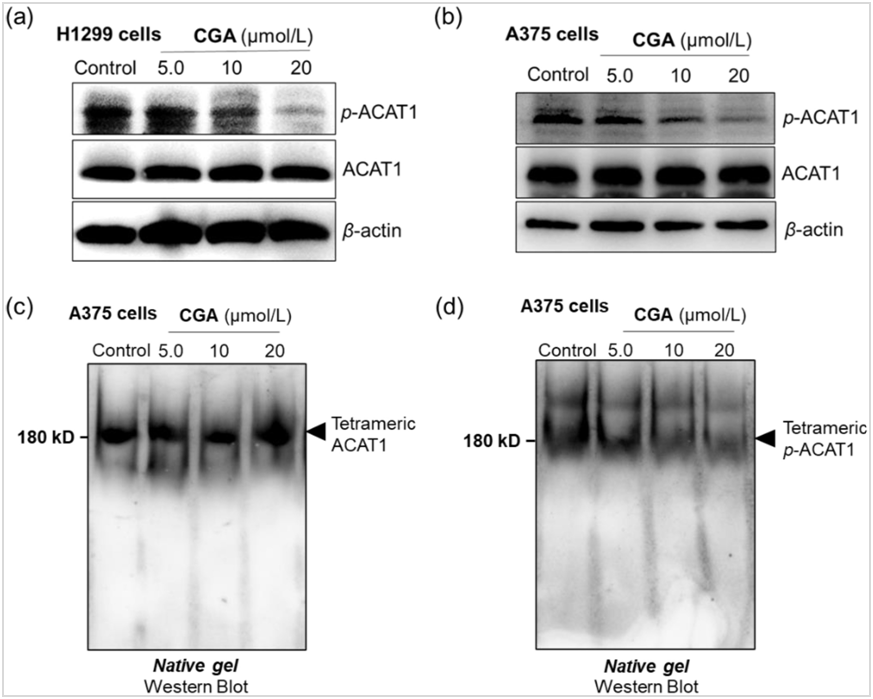
Figure 5. CGA is a no-covalent ACAT1 Inhibitor, disrupting active ACAT1 function by eliminating Y407 phosphorylation without hindering tetramer reformation.
(a, b) Western blot assay of phospho-ACAT1(Y407) and ACAT1 expression in the A375 cells and NCI-H1299 cells; (c, d) Western blot assay of tetramer ACAT1/Phospho- ACAT1(Y407) proteins in the A375 cells treated with or without CGA by native page.
The identification that ACAT1 was the target or substrate of CGA allowed us to further evaluate how CGA influenced ACAT1 activity. To that end, in vitro CGA treatment experiments on A375 cells and NCI-H1299 cells were performed. We observed that CGA treatment effectively inhibited Y407 phosphorylation in both A375 and NCI-H1299 cell lines, consistent with its effects in glioblastoma cells (Fig. 5a, b and Supporting Information Fig. S18), and significantly increased PDH activity in A375 cells compared to control untreated cells (Supporting Information Fig. S19). Furthermore, CGA treatment in A375 cell lines did not influence tetramer formation but decreased Y407 phosphorylation of tetramer ACAT1 on a native gel electrophoresis (Fig. 5c and d). These results suggested that the apparent inhibition of ACAT1 by CGA might be due to the interruption of Y407 phosphorylation, rather than disrupting ACAT1 tetramers into inactive monomers.
Next, in vivo CGA treatments were carried out to validate its mode of action in xenograft nude mice with subcutaneous injection of A375 cells or A375 cells with ACAT1 knockdown (A375 shACAT1). In the xenograft experiments, the growth rate and masses (Fig. 6a‒c, Supporting Information Fig. S20) of tumors derived from A375 shACAT1 cells were significantly reduced compared with control cells expressing wildtype ACAT1 (ACAT1 WT). This reduction is attributed to diminished ACAT1 activity, leading to decreased cancer cell proliferation.
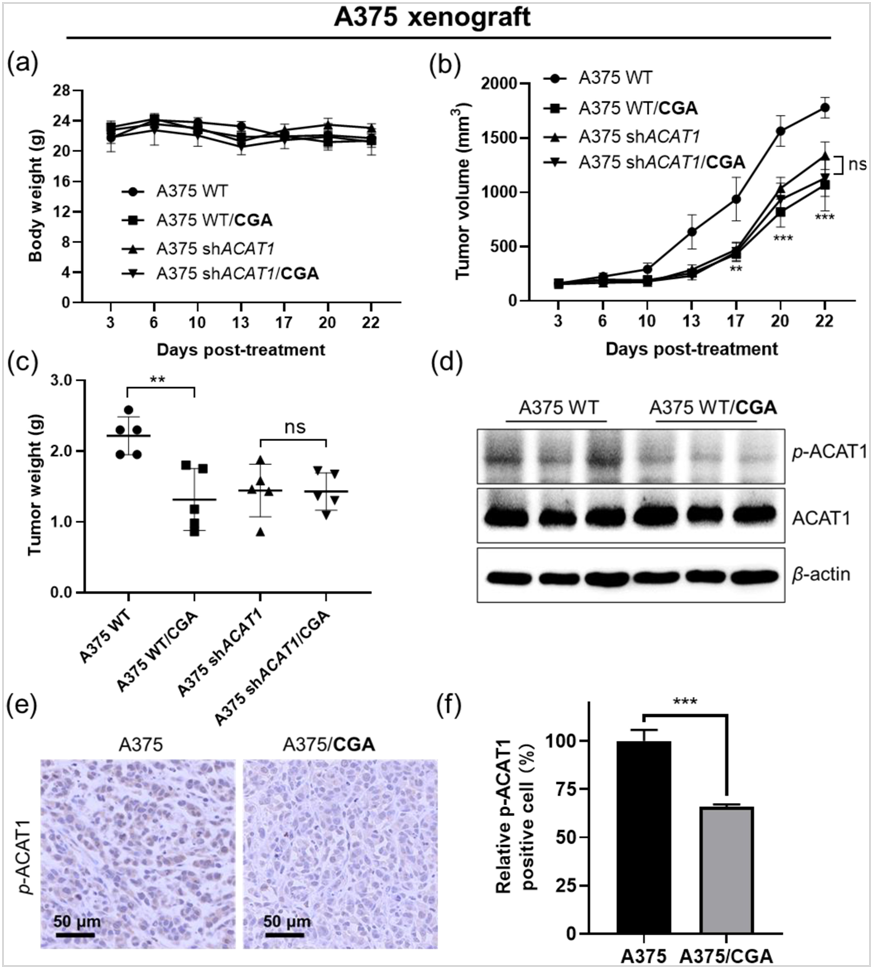
Figure 6 CGA significantly reduces Y407 phosphorylation, inhibiting cancer cell proliferation and tumor growth.
(a‒c) Effects of CGA administered via intraperitoneal injection in A375 xenograft nude mice on body weight, tumor growth rate and mass; (d) Western blot assay of phospho-ACAT1(Y407) expression in the A375 tumor; (e) Phospho-ACAT1(Y407) immunohistochemical staining of A375 tumor. Scale bar, 50 μmol/L; (f) Quantification of intensities from phospho-ACAT1(Y407) immunohistochemical staining, *P< 0.05, **P < 0.01 and ***P < 0.001. Data present as mean±SD.
Furthermore, A375 xenografts under CGA treatment (50 mg/kg/day) showed reduced tumor growth compared to non-treated control mice, with no effect on body weight in drug-treated nude mice. Interestingly, tumor volume and weight of A375 shACAT1 xenograft in mice were not furtherly reduced when treated with CGA compared to control (A375 shACAT1). A375 xenograft mice with CGA treatment decreased ACAT1 activity in resected tumors from xenograft nude compared to control without CGA treatment (Fig. 6d‒f). Similar results were obtained in the models of NCI-H1299 and ACAT1 knockdown NCI-H1299 shACAT1 xenograft nude mice, which were treated with CGA for 21 days (Supporting Information Figs. S21 and S22). Together, these data suggested CGA was a novel non-covalent ACAT1 inhibitor that efficiently bound to ACAT1 tetramers, abolished Y407 phosphorylation, and then decreased ACAT1 activity via a new mode of action, which removed the inhibitory effect of ACAT1 on PDH, attenuated cancer cell proliferation and tumor growth.
4. Conclusions
Natural products have been major sources of therapeutic agents to treat various diseases in human history. However, targets or molecular mechanisms of action remain unidentified for substantial numbers of natural products. In this study, using a novel bifunctional photo-affinity probe PAL/CGA and AfBPP chemical proteomic approach, we discover, for the first time, the mitochondrial ACAT1 is one of the main target proteins of CGA. The binding affinity for CGA and ACAT1 was confirmed by multiple assays, including SPR, ITC, TSA, and DARTS, using purified recombinant ACAT1 protein and cell lysates. Most importantly, the first cryo-EM structure of tetrameric ACAT1 in complex with CGA was obtained and revealed informative details for CGA binding sites and the mechanism of inhibition of ACAT1. Contrary to the reported covalent inhibit AH which inhibits ACAT1 activity by disrupting ACAT1 tetramers into inactive monomers, there is no impact by CGA on the equilibrium between tetrameric and monomeric ACAT1 proteins. Systematic in vitro, cellular, and in vivo experiments have provided a clear view of the mode of action of CGA at the molecular level. CGA non-covalently binds to ACAT1 tetramers, induces an extensive conformational change, leading to abolishing Y407 phosphorylation and then decreasing ACAT1 activity. ACAT1, a key metabolic enzyme, not only catalyzes the transformation of two acetyl-CoA molecules into acetoacetyl-CoA and CoA but also functions as an acetyl transferase for the pyruvate dehydrogenase complex. This establishes ACAT1 as a crucial regulatory point between glycolysis and oxidative phosphorylation, influencing the metabolic pathway choice at acetoacetyl-CoA44. While dysfunctional ACTA1 is important in regulating cancer cell’s metabolism, currently there is no FDA-approved drug for this target. This new mode of action by CGA on ACAT1 in turn removes the inhibitory effect of ACAT1 on PDH, attenuates cancer cell proliferation and tumor growth.
Thus, our studies shed light on the application of AfBPP platforms in target fishing and protein identification, and as a novel chemical tool to explore ACAT1 biological functions. Importantly, the identificaton of ACAT1 as one of the main targets for CGA helps to explain the clinical benefit conferred by CGA, the widely existing natural product and warrants future studies to use ACAT1 as a potential biomarker for clinical applications.
Acknowledgments
CGA was kindly provided by Sichuan Jiuzhang Biological Science and Technology Co., Ltd. We acknowledge Dr. Kehui Zhang (IMM) and Dr. Xiaoyu Wang (IMM) for their help with the creation of target mapping experiments. We thank Shuimu BioSciences for Cryo-EM image acquisitions and the assistance of data analysis, especially Dr. Chuan Liu, Dr. Jing Li, Dr. Fanhao Meng and Dr. Xiaodan Ni. We alsothank Dr. Jing Deng from Sichuan Jiuzhang Biological Science and Technology Co., Ltd. for helpful discussions and critical reading of the manuscript. The work was partially supported and inspired by the National Natural Science Foundation of China (NSFC) projects (22122705, 22077139, 82293684) and CAMS Innovation Fund fo Medical Sciences (CIFMS) (2021-I2M-1-054, 2022-I2M-2-002, 2022-I2M-1-014, China).
Author contributions
Hai-Yu Hu and Xiaoguang Chen contributed to the central experimental idea. Qinghua Wang, Tingting Du, Zhihui Zhang, Qingyang Zhang, Jie Zhang, and Wenbin Lin designed and performed the experiments. Qinghua Wang wrote the original draft. Jian-Dong Jiang, Xiaoguang Chen and Hai-Yu Hu supervised the project, revised the
manuscript and commented on it. All authors have approved the final version of the manuscript.
Conflicts of interest
Jie Zhang is a shareholder in Sichuan Jiuzhang Biological Science and Technology Co., Ltd. All other authors declare no competing interests.
References
1. Clifford MN, Jaganath IB, Ludwig IA, Crozier A. Chlorogenic acids and the acylquinic acids: discovery, biosynthesis, bioavailability and bioactivity. Nat Prod Rep 2017;34:1391−421.
2. Lu HJ, Tian ZM, Cui YY, Liu ZC, Ma XY. Chlorogenic acid: a comprehensive review of the dietary sources, processing effects, bioavailability, beneficial properties, mechanisms of action, and future directions. Compr Rev Food Sci F 2020;19:3130−58.
3. Machado F, Coimbra MA, del Castillo MD, Coreta-Gomes F. Mechanisms of action of coffee bioactive compounds‒a key to unveil the coffee paradox. Crit Rev Food Sci 2023, 20:1−23.
4. Caruso G, Torrisi SA, Mogavero MP, Currenti W, Castellano S, Godos J, et al. Polyphenols and neuroprotection: therapeutic implications for cognitive decline. Pharmacol Therapeut 2022;232:108013.
5. Xue HY, Wei MJ, Ji LL. Chlorogenic acids: a pharmacological systematic review on their hepatoprotective effects. Phytomedicine 2023;118:154961.
6. Kang Z, Li S, Kang X, Deng J, Yang HR, Chen F, et al. Phase I study of chlorogenic acid injection for recurrent high-grade glioma with long-term follow-up. Cancer Biol Med 2023;20:465−76.
7. Gupta A, Atanasov AG, Li YT, Kumar N, Bishayee A. Chlorogenic acid for cancer prevention and therapy: current status on efficacy and mechanisms of action. Pharmacol Res 2022;186:106505.
8. Huang S, Wang LL, Xue NN, Li C, Guo HH, Ren TK, et al. Chlorogenic acid effectively treats cancers through induction of cancer cell differentiation. Theranostics 2019;9: 6745−6763.
9. Zheng C, Zhong Y, Zhang W, Wang Z, Xiao H, Zhang W, et al. Chlorogenic acid ameliorates post-infectious irritable bowel syndrome by regulating extracellular vesicles of gut microbes. Adv Sci (Weinh) 2023;10:e2302798.
10. Ke Y, Ma Z, Ye H, Guan X, Xiang Z, Xia Y, et al. Chlorogenic acid-conjugated nanoparticles suppression of platelet activation and disruption to tumor vascular barriers for enhancing drug penetration in tumor. Adv Healthc Mater 2023;12:e2202205.
11. Cao H, Yang L, Tian R, Wu H, Gu Z, Li Y. Versatile polyphenolic platforms in regulating cell biology. Chem Soc Rev 2022;51:4175−4198.
12. Rhee KY, Jansen RS, Grundner C. Activity-based annotation: the emergence of systems biochemistry. Trends Biochem Sci 2022;47:785−794.
13. Spradlin JN, Zhang E, Nomura DK. Reimagining druggability using chemoproteomic platforms. Acc Chem Res 2021;54:1801−1813.
14. Parthasarathy A, Mantravadi PK, Kalesh K. Detectives and helpers: natural products as resources for chemical probes and compound libraries. Pharmacol Therapeut 2020;216: 107688.
15. Spradlin JN, Hu XR, Ward CC, Brittain SM, Jones MD, Ou LS, et al. Harnessing the anti-cancer natural product nimbolide for targeted protein degradation. Nat Chem Biol 2019;15:747−755.
16. McGregor NGS, de Boer C, Foucart QPO, Beenakker T, Offen WA, Codée JDC, et al. A multiplexing activity-based protein-profiling platform for dissection of a native bacterial xyloglucan-degrading system. Acs Central Sci 2023;9:2306−2314.
17. Meissner F, Geddes-McAlister J, Mann M, Bantscheff M. The emerging role of mass spectrometry-based proteomics in drug discovery. Nat Rev Drug Discov 2022;21:637−54.
18. Chen X, Wang Y, Ma N, Tian J, Shao Y, Zhu B, et al. Target identification of natural medicine with chemical proteomics approach: probe synthesis, target fishing and protein identification. Signal Transduct Target Ther 2020;5:72.
19. Böttcher T, Pitscheider M, Sieber SA. Natural products and their biological targets: proteomic and metabolomic labeling strategies. Angew Chem Int Edit 2010;49:2680−98.
20. Fang HX, Peng B, Ong SY, Wu Q, Li L, Yao SQ. Recent advances in activity-based probes (ABPs) and affinity-based probes (ABPs) for profiling of enzymes. Chem Sci2021;12:8288−310.
21. Korovesis D, Beard HA, Mérillat C, Verhelst SHL. Probes for photoaffinity labelling of kinases. Chembiochem 2021;22: 2206−18.
22. Hsu KL. Shining a light on phenotypic drug discovery. Cell Chem Biol 2021;28:115−117.
23. Grammel M, Hang HC. Chemical reporters for biological discovery. Nat Chem Biol 2013;9:475−484.
24. Zhang KH, Sun W, Huang LH, Zhu KY, Pei F, Zhu LC, et al. Identifying glyceraldehyde 3-phosphate dehydrogenase as a cyclic adenosine diphosphoribose binding protein by photoaffinity protein‒ligand labeling approach. J Am Chem Soc 2017;139:156−170.
25. Blagg J, Workman P. Choose and use your chemical probe wisely to explore cancer biology. Cancer Cell 2017;32:9−25.
26. Wright MH, Sieber SA. Chemical proteomics approaches for identifying the cellular targets of natural products. Nat Prod Rep 2016;33:681−708.
27. Joneidi S, Alizadeh SR, Ebrahimzadeh MA. Chlorogenic acid derivatives: structural modifications, drug design, and biological activities: a review. Mini Rev Med Chem 2024; 24:748−66.
28. Sehrawat R, Rathee P, Rathee P, Khatkar S, Akkol EK, Khatkar A, et al. In silico design of novel bioactive molecules to treat breast cancer with chlorogenic acid derivatives: a computational and SAR approach. Front Pharmacol 2023;14:1266833.
29. Pressete CG, Viegas FPD, Campos TG, Caixeta ES, Hanemann JAC, Ferreira-Silva GA, et al. Piperine-chlorogenic acid hybrid inhibits the proliferation of the SK-MEL-147 melanoma cells by modulating mitotic kinases. Pharmaceuticals 2023;16:145.
30. Pan SJ, Zhang HL, Wang CY, Yao SCL, Yao SQ. Target identification of natural products and bioactive compounds using affinity-based probes. Nat Prod Rep 2016;33:612−620.
31. Tao YF, Felber JG, Zou ZY, Njomen E, Remsberg JR, Ogasawara D, et al. Chemical proteomic discovery of isotype-selective covalent inhibitors of the RNA methyltransferase NSUN2. Angew Chem Int Edit 2023; 62: e202311924.
32. Lazear MR, Remsberg JR, Jaeger MG, Rothamel K, Her HL, DeMeester KE, et al. Proteomic discovery of chemical probes that perturb protein complexes in human cells. Mol Cell 2023;83:1725−1742.
33. Wang QH, Zhang QY, Zhang ZH, Ji M, Du TT, Jin J, et al. Characterization of chlorogenic acid as a two-photon fluorogenic probe that regulates glycolysis in tumor cells under hypoxia. J Med Chem 2023;66:2498−2505.
34. Fan J, Lin RT, Xia SY, Chen D, Elf SE, Liu SP, et al. Tetrameric acetyl-CoA acetyltransferase 1 is important for tumor growth. Mol Cell 2016;64:859−74.
35. Pavlova NN, Zhu JJ, Thompson CB. The hallmarks of cancer metabolism: still emerging. Cell Metab 2022;34:355−77.
36. Finley LWS. What is cancer metabolism?. Cell 2023;186:1670−88.
37. Stine ZE, Schug ZT, Salvino JM, Dang CV. Targeting cancer metabolism in the era of precision oncology. Nat Revs Drug Discov 2022;21:141−62.
38. Xiao Y, Yu TJ, Xu Y, Ding R, Wang YP, Jiang YZ, et al. Emerging therapies in cancer metabolism. Cell Metab 2023;35:1283−303.
39. Goudarzi A. The recent insights into the function of ACAT1: a possible anti-cancer therapeutic target. Life Sci 2019;232: 116592.
40. Lo YW, Lin ST, Chang SJ, Chan CH, Lyu KW, Chang JF, et al. Mitochondrial proteomics with siRNA knockdown to reveal ACAT1 and MDH2 in the development of doxorubicin-resistant uterine cancer. J Cell Mol Med 2015;19:744−59.
41. Fan J, Shan CL, Kang HB, Elf S, Xie JX, Tucker M, et al. Tyr phosphorylation of PDP1 toggles recruitment between ACAT1 and SIRT3 to regulate the pyruvate dehydrogenase complex. Mol Cell 2014;53:534−48.
42. Gu L, Zhu YH, Lin X, Tan XY, Lu BJ, Li YJ. Stabilization of FASN by ACAT1-mediated GNPAT acetylation promotes lipid metabolism and hepatocarcinogenesis. Oncogene 2020;39:2437−49.
43. Wang M, Wang W, You S, Hou Z, Ji M, Xue N, et al. ACAT1 deficiency in myeloid cells promotes glioblastoma progression by enhancing the accumulation of myeloid-derived suppressor cells. Acta Pharm Sin B 2023;13:4733−47.
44. Garcia-Bermudez J, Birsoy K. Drugging ACAT1 for cancer therapy. Mol Cell 2016;64:856−7.







